NEBEspresso¶
Derived from the ase.neb.NEB class, NEBEspresso orchestrates (i)Espresso calculators of individual images in a NEB calculation. This way a NEBEspresso object facilitates parallel NEB jobs, where each image calculator uses a subset of the pool of CPUs available to the job.
This tutorial explains the use of NEBEspresso to do a NEB calculation on methyl group rotation in ethane, built on this ASE example.
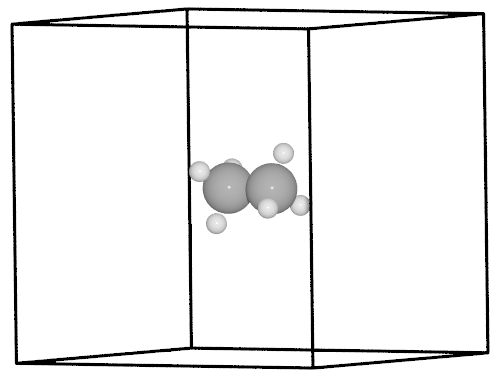
Initial and final structures of the NEB¶
As opposed to the Effective Medium Potential used in the original example, Quantum Espresso needs periodic boundaries defined by a unit cell. That is defined in the ethane.traj trajectory file containing the optimized structure. The final structure is created simply by permuting the H atoms in one of the methyl groups.
from espresso import iEspresso
from ase.io import read
initial = read('ethane.traj')
initial.get_potential_energy()
final = initial.copy()
final.positions[2:5] = initial.positions[[3, 4, 2]]
final.get_potential_energy()
Assigning calculators to intermediate images¶
Initially, images are created as copies of the initial structure and are each asssigned a calculator. Here we use the interactive iEspresso calculator, but the non-interactive Espresso calculator is equally valid.
images = [initial]
for _ in range(7):
image = initial.copy()
image.set_calculator(iEspresso(pw=300, dw=4000,kpts='gamma',xc='PBE'))
images.append(image)
images.append(final)
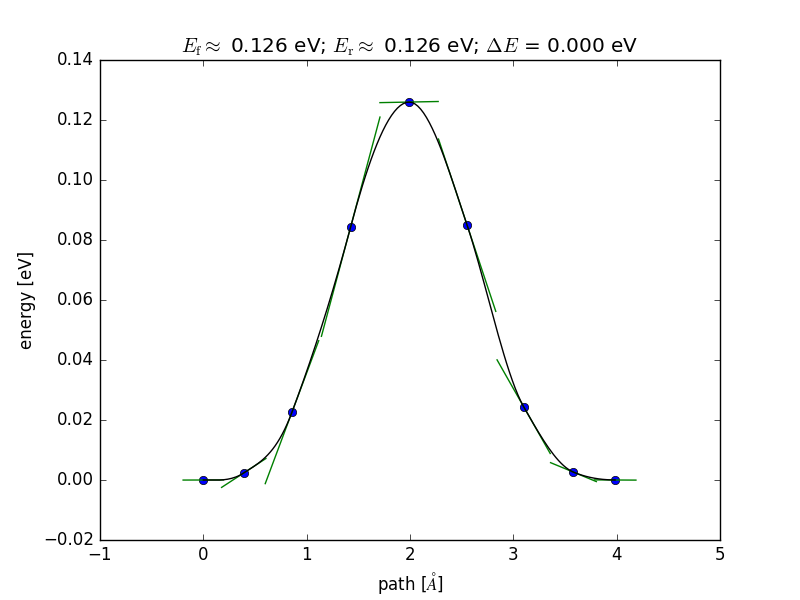
Running the NEB calculation and analyzing the results¶
The NEBEspresso class is instantiated with the list of images and is used just as the ase.neb.NEB super class, with one exception: the parallel keyword. NEBEspresso ignores 'parallel'=False and will always attempt to distribute the image calculators over the CPU pool available to the job.
from espresso.nebespresso import NEBEspresso
from ase.optimize.fire import FIRE as QuasiNewton
neb = NEBEspresso(images)
neb.interpolate('idpp')
qn = QuasiNewton(neb, logfile='ethane_linear.log', trajectory='neb.traj')
qn.run(fmax=0.05)
from ase.neb import NEBTools
nt = NEBTools(neb.images)
fig = nt.plot_band()
fig.savefig('rotation-barrier.png')